Abstract
Research Article
Characterization of Salmonella spp. isolated from small turtles and human in Republic of Korea
Su-Jin Chae, Jin-Suk Lim and Deog-Yong Lee*
Published: 11 December, 2020 | Volume 4 - Issue 1 | Pages: 051-055
In 2013, the World Health Organization (WHO) reported that small, pet turtles had caused multistate Salmonella outbreaks in the United States, from where small turtles were subsequently exported into the Republic of Korea. We investigated cases of salmonellosis in South Korea associated with domestic small turtles and analysed genetic characteristics of Salmonella isolates in commercially-available small turtles. We traced six Salmonella serovars, known to have caused human infection in the United States (S. Sandiego, S. Pomona, S. Poona, S. Newport, I 4,(5),12:i:-, and S. Typhimurium), in isolates from suspected Salmonella infection cases in Korea from 2006 to 2015. Additionally, we conducted a pilot study of isolates from small turtles being sold in Korean markets, and performed molecular genetic analysis on the identified strains. S. Pomona was identified in one Salmonella infection case, while all strains isolated from small turtles belonged to either subspecies I (enterica, n = 10, 71.4%) or subspecies IIIb (diarizonae, n = 4, 28.6%). Two serovars (S. Pomona and S. Sandiego) that were highly associated with turtle-to-human transmission were identified with 100% homology to human isolates. Previous to this study, turtle-associated human S. Pomona infections were not well reported in Korea. We report Salmonella infection in small turtles in Korea, and confirm that small turtles should be considered the first infectious agent in S. Pomona infection. We therefore suggest quarantine measures for importing small turtles be enhanced in Korea.
Read Full Article HTML DOI: 10.29328/journal.ivs.1001027 Cite this Article Read Full Article PDF
Keywords:
Salmonella infection; Turtles; Zoonoses; Multilocus sequence typing (MLST); Pulsed-field gel electrophoresis (PFGE)
References
- Salmonellosis associated with pet turtles--Wisconsin and Wyoming, 2004. MMWR. 2005; 54. 223-226. PubMed: https://pubmed.ncbi.nlm.nih.gov/15758895/
- Seepersadsingh N, Adesiyun AA. Prevalence and antimicrobial resistance of Salmonella spp. in pet mammals, reptiles, fish aquarium water, and birds in Trinidad. J Vet Med B Infect Dis Vet Public Health. 2003; 50: 488-493. PubMed: https://pubmed.ncbi.nlm.nih.gov/14720186/
- Woodward DL, Khakhria R, Johnson WM. Human salmonellosis associated with exotic pets. J Clin Microbiol. 1997; 35: 2786-2790. PubMed: https://pubmed.ncbi.nlm.nih.gov/9350734/
- DuPonte MW, Nakamura RM, Chang EM. Activation of latent Salmonella and Arizona organisms by dehydration of red-eared turtles, Pseudemys scripta-elegans. Am J Vet Re. 1978; 39: 529-530. PubMed: https://pubmed.ncbi.nlm.nih.gov/637401/
- Bosch S, Tauxe RV, Behravesh CB. Turtle-Associated Salmonellosis, United States, 2006-2014. Emerg Infect Dis. 2016; 22: 1149-1155. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4918145/
- Lee DY, Lee E, Min JE, Kim SH, Oh HB, et al. Epidemic by Salmonella I 4,[5], 12: i:-and characteristics of isolates in Korea. J Infection Chemotherapy. 2011; 43:186-190.
- Kim SH, Kim SH, Lee SW, Kang YH, Lee BK. Rapid serological identification for monophasic Salmonella serovars with a hin gene: specific polymerase chain reaction. J Bacteriol Virol. 2005; 35: 291-298.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual: Cold spring harbor laboratory press; 1989.
- Grimont PAD, Weill FX. Antigenic formulae of the Salmonella serovars. WHO collaborating centre for reference on Salmonella. 2007; 9: 1-166.
- Issenhuth-Jeanjean S, Roggentin P, Mikoleit M, Guibourdenche M, de Pinna E, et al. Supplement 2008–2010 (no. 48) to the white–Kauffmann–Le minor scheme. J Research in microbiology. 2014; 165: 526-530. PubMed: https://pubmed.ncbi.nlm.nih.gov/25049166/
- Yun YS, Chae SJ, Na HY, Chung GT, Yoo CK, et al. Modified method of multilocus sequence typing (MLST) for serotyping in Salmonella species. J Bacteriol Virol. 2015; 45: 314-318.
- Institute CaLS. Performance standards for antimicrobial susceptibility testing. 2018; 23.
- Geue L, Loschner U. Salmonella enterica in reptiles of German and Austrian origin. Vet Microbiol. 2002; 84: 79-91. PubMed: https://pubmed.ncbi.nlm.nih.gov/11731161/
- Meyer Sauteur PM, Relly C, Hug M, Wittenbrink MM, Berger C. Risk factors for invAsive reptile-associated salmonellosis in children. Vector Borne Zoonotic Dis. 2013; 13: 419-421. https://pubmed.ncbi.nlm.nih.gov/23473215/
- Nakadai A, Kuroki T, Kato Y, Suzuki R, Yamai S, Yaginuma C, et al. Prevalence of Salmonella spp. in pet reptiles in Japan. J Vet Med Sci. 2005; 67: 97-101. PubMed: https://pubmed.ncbi.nlm.nih.gov/15699603/
- Khuly P, Meyer T, Carlson R, Delamaide Gasper JA, Rylander H, et al. Eight multistate outbreaks of human Salmonella infections linked to small turtles (final update). J Am Vet Med Assoc. 2014; 244: 1243-1245.
- Trade statics system. Korea Custom Service. 2018. http://www.customs.go.kr.
- Kim JD, Choi SY, Kim DS, Kim KH. Two Children with Nontyphoidal Salmonellosis Assumed by Pets Korean J Pediatr Infect Dis. 2013; 20: 41-45.
- Chae SJ, Yun YS, Yoo CK, Chung GT, Lee DY. First Report of Salmonella Serotype Tilene Infection in Korea. J Annals of Clinical Microbiology. 2016; 19: 24-27.
- Lee DY, Kang Y. Human infection of Salmonella matadi in Korea. Yonsei Med J. 2013; 54: 1297-1298. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3743207/
- Turtle-associated salmonellosis in humans--United States, 2006-2007. MMWR. 2007; 56: 649.
- Tauxe RV, Rigau-Perez JG, Wells JG, Blake PA. Turtle-associated salmonellosis in Puerto Rico. Hazards of the global turtle trade. JAMA. 1985; 254: 237-239. PubMed: https://pubmed.ncbi.nlm.nih.gov/3999366/
- Cooke FJ, De Pinna E, Maguire C, Guha S, Pickard DJ, et al. First report of human infection with Salmonella enterica serovar Apapa resulting from exposure to a pet lizard. J Clin Microbiol. 2009; 47: 2672-2674. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5617995/
- Chen CY, Chen WC, Chin SC, Lai YH, Tung KC, et al. Prevalence and antimicrobial susceptibility of Salmonellae isolates from reptiles in Taiwan. J Vet Diagn Invest. 2010; 22: 44-50. PubMed: https://pubmed.ncbi.nlm.nih.gov/20093681/
Figures:
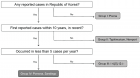
Figure 1
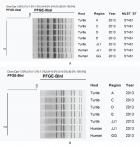
Figure 2

Figure 3
Similar Articles
-
Characterization of Salmonella spp. isolated from small turtles and human in Republic of KoreaSu-Jin Chae,Jin-Suk Lim,Deog-Yong Lee*. Characterization of Salmonella spp. isolated from small turtles and human in Republic of Korea. . 2020 doi: 10.29328/journal.ivs.1001027; 4: 051-055
-
Fish-borne parasites proficient in zoonotic diseases: a mini reviewAvishek Bardhan*. Fish-borne parasites proficient in zoonotic diseases: a mini review. . 2022 doi: 10.29328/journal.ivs.1001035; 6: 005-012
Recently Viewed
-
Transumbilical Single-incision Hiatal Hernia Repair and Nissen Fundoplication in situs Inversus Totalis: A Rare Case ReportQing Cao,Chen Kang,Kang Gu,Yin Peng,Yang Lv,Xu-Zhong Ding,Peng Li*. Transumbilical Single-incision Hiatal Hernia Repair and Nissen Fundoplication in situs Inversus Totalis: A Rare Case Report. Adv Treat ENT Disord. 2026: doi: 10.29328/journal.ated.1001017; 10: 001-003
-
NAD⁺ Biology in Ageing and Chronic Disease: Mechanisms and Evidence across Skin, Fertility, Osteoarthritis, Hearing and Vision Loss, Gut Health, Cardiovascular–Hepatic Metabolism, Neurological Disorders, and MuscleRizwan Uppal,Umar Saeed*,Muhammad Rehan Uppal. NAD⁺ Biology in Ageing and Chronic Disease: Mechanisms and Evidence across Skin, Fertility, Osteoarthritis, Hearing and Vision Loss, Gut Health, Cardiovascular–Hepatic Metabolism, Neurological Disorders, and Muscle. Ann Clin Endocrinol Metabol. 2026: doi: 10.29328/journal.acem.1001032; 10: 001-009
-
Treatment Outcome in Patients with Myofascial Orofacial Pain: A Randomized Clinical TrialAnders Wänman*, Susanna Marklund, Negin Yekkalam. Treatment Outcome in Patients with Myofascial Orofacial Pain: A Randomized Clinical Trial. J Oral Health Craniofac Sci. 2024: doi: 10.29328/journal.johcs.1001046; 9: 001-008
-
Anxiety and depression as an effect of birth order or being an only child: Results of an internet survey in Poland and GermanyJochen Hardt*,Lisa Weyer,Malgorzata Dragan,Wilfried Laubach. Anxiety and depression as an effect of birth order or being an only child: Results of an internet survey in Poland and Germany. Insights Depress Anxiety. 2017: doi: 10.29328/journal.hda.1001003; 1: 015-022
-
Case Report of a Child with Beta Thalassemia Major in a Tribal Region of IndiaNeha Chauhan, Prakash Narayan, Mahesh Narayan, Manisha Shukla*. Case Report of a Child with Beta Thalassemia Major in a Tribal Region of India. J Child Adult Vaccines Immunol. 2023: doi: 10.29328/journal.jcavi.1001011; 7: 005-007
Most Viewed
-
Impact of Latex Sensitization on Asthma and Rhinitis Progression: A Study at Abidjan-Cocody University Hospital - Côte d’Ivoire (Progression of Asthma and Rhinitis related to Latex Sensitization)Dasse Sery Romuald*, KL Siransy, N Koffi, RO Yeboah, EK Nguessan, HA Adou, VP Goran-Kouacou, AU Assi, JY Seri, S Moussa, D Oura, CL Memel, H Koya, E Atoukoula. Impact of Latex Sensitization on Asthma and Rhinitis Progression: A Study at Abidjan-Cocody University Hospital - Côte d’Ivoire (Progression of Asthma and Rhinitis related to Latex Sensitization). Arch Asthma Allergy Immunol. 2024 doi: 10.29328/journal.aaai.1001035; 8: 007-012
-
Causal Link between Human Blood Metabolites and Asthma: An Investigation Using Mendelian RandomizationYong-Qing Zhu, Xiao-Yan Meng, Jing-Hua Yang*. Causal Link between Human Blood Metabolites and Asthma: An Investigation Using Mendelian Randomization. Arch Asthma Allergy Immunol. 2023 doi: 10.29328/journal.aaai.1001032; 7: 012-022
-
An algorithm to safely manage oral food challenge in an office-based setting for children with multiple food allergiesNathalie Cottel,Aïcha Dieme,Véronique Orcel,Yannick Chantran,Mélisande Bourgoin-Heck,Jocelyne Just. An algorithm to safely manage oral food challenge in an office-based setting for children with multiple food allergies. Arch Asthma Allergy Immunol. 2021 doi: 10.29328/journal.aaai.1001027; 5: 030-037
-
Snow white: an allergic girl?Oreste Vittore Brenna*. Snow white: an allergic girl?. Arch Asthma Allergy Immunol. 2022 doi: 10.29328/journal.aaai.1001029; 6: 001-002
-
Cytokine intoxication as a model of cell apoptosis and predict of schizophrenia - like affective disordersElena Viktorovna Drozdova*. Cytokine intoxication as a model of cell apoptosis and predict of schizophrenia - like affective disorders. Arch Asthma Allergy Immunol. 2021 doi: 10.29328/journal.aaai.1001028; 5: 038-040

If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."